CellPose
Description
A generalist algorithm for cellular segmentation. Cellpose-SAM: cell and nucleus segmentation with superhuman generalization. It can be optimized for your own data, applied in 3D, works on images with shot noise, (an)isotropic blur, undersampling, contrast inversions, regardless of channel order and object sizes.
Keywords
SAM, deep learning, nucleus segmentation
Supported Versions
- 3.1.1.1
- 4.0.8 (CellPose-SAM)
Documentation
Example Usage
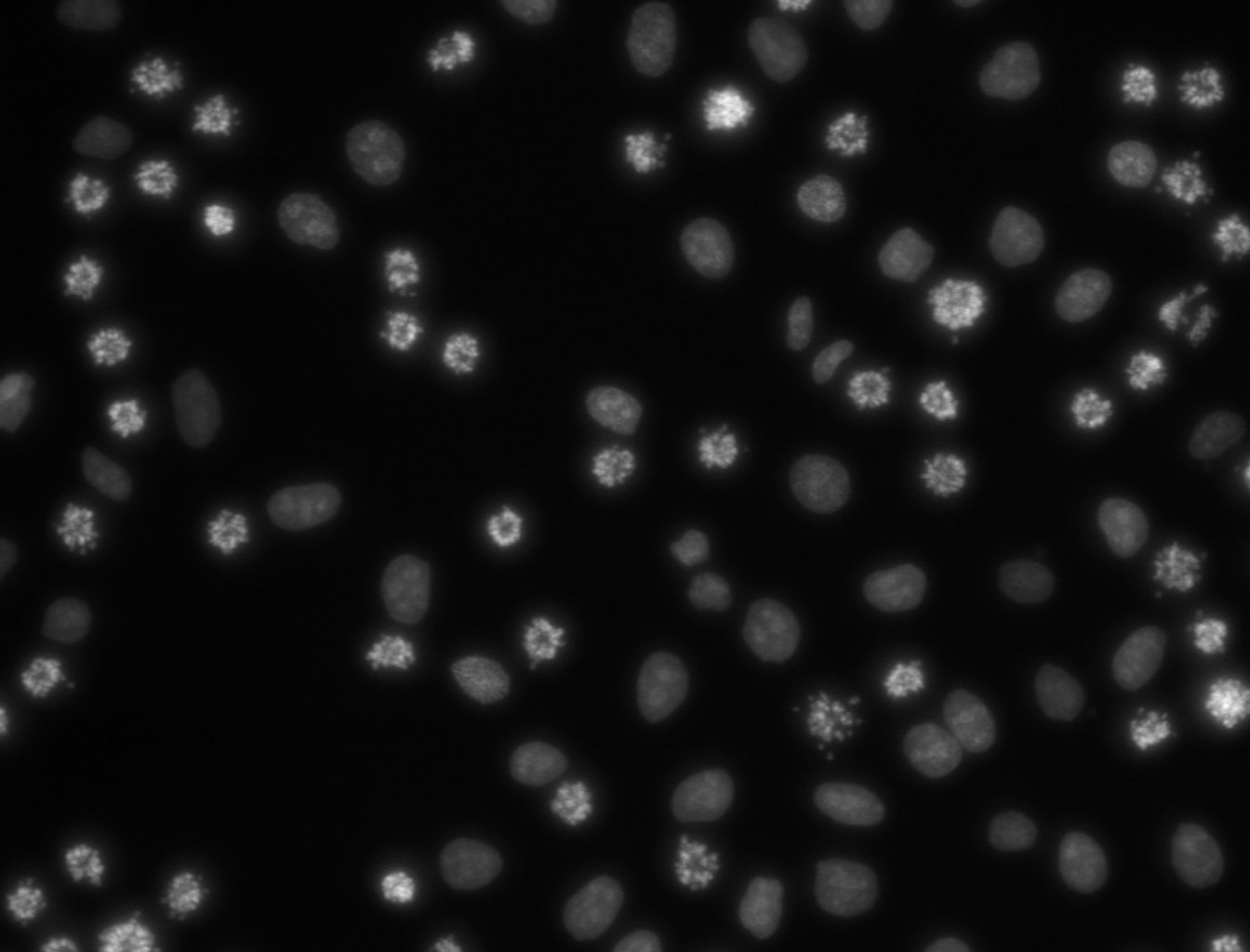
Original
⟹

Mask
The image on the left shows a population of cells, including several in apoptotic states. Segmentation was performed using CellPose-SAM, with the SAM model. The image on the right is the resulting mask.
Python
!pip install git+https://www.github.com/mouseland/cellpose.git
import numpy as np
from cellpose import models, core, io, plot
from pathlib import Path
from tqdm import trange
import matplotlib.pyplot as plt
from natsort import natsorted
io.logger_setup() # run this to get printing of progress
#Check if notebook instance has GPU access
if core.use_gpu()==False:
raise ImportError("No GPU access, change your runtime")
model = models.CellposeModel(gpu=True)
# segment 1 image in folder
img = io.imread("./2d_cells_apoptotic_1channel.jpg")
first_channel = '0' # @param ['None', 0, 1, 2, 3, 4, 5]
second_channel = '1' # @param ['None', 0, 1, 2, 3, 4, 5]
third_channel = '2' # @param ['None', 0, 1, 2, 3, 4, 5]
selected_channels = []
for i, c in enumerate([first_channel, second_channel, third_channel]):
if c == 'None':
continue
if int(c) > img.shape[-1]:
assert False, 'invalid channel index, must have index greater or equal to the number of channels'
if c != 'None':
selected_channels.append(int(c))
img_selected_channels = np.zeros_like(img)
img_selected_channels[:, :, :len(selected_channels)] = img[:, :, selected_channels]
flow_threshold = 0.4
cellprob_threshold = 0.0
tile_norm_blocksize = 0
masks, flows, styles = model.eval(img_selected_channels, batch_size=32, flow_threshold=flow_threshold, cellprob_threshold=cellprob_threshold,
normalize={"tile_norm_blocksize": tile_norm_blocksize})
fig = plt.figure(figsize=(12,5))
plot.show_segmentation(fig, img_selected_channels, masks, flows[0])
plt.tight_layout()
plt.show()